Hijacking of plasmin by dengue virus for infection
February 03, 2025Biological scientists from the National University of Singapore (NUS) have uncovered how the dengue virus uses its envelope protein to capture human plasmin from a blood meal to enhance the permeability of the mosquito midgut for infection.
Plasmin is a protease that contains five kringle-domains (KR1-5) responsible for substrate binding. In addition to digesting blood clots, plasmin is also used for the breakdown of the extracellular matrix to enable cell movement in tissues. While bacteria are known to capture human plasmin to digest host tissue for metastasis, the hijacking of plasmin by viruses for infection is not well studied.
The dengue virus is transmitted to humans through mosquito bites. Once ingested in a blood meal, the virus needs to pass through the mosquito midgut to infect the salivary glands before being transmitted to the next human host. However, the mechanism enabling this midgut traversal is not well understood. The acidic motifs on both the KR-4 and KR-5 domains of plasmin are found to bind synergistically to two lysine-containing regions (basic) located on the domain I of the dengue virus envelope protein. This interaction enhances the permeability of the mosquito midgut, facilitating viral infection. The identification of the exact binding sites between plasmin and dengue virus provides a potential way to interfere with this interaction and prevent dengue virus transmission.
A research team led by Associate Professor MOK Yu Keung from the Department of Biological Science at NUS expressed individual domains of human plasmin and the dengue virus envelope protein using an insect cell system. Kinetic binding experiments show that both KR-4 and KR-5 domains are needed to bind synergistically to the dengue virus. In addition, hydrogen-deuterium mass spectrometry experiments showed that two lysine-containing regions on domain I of the dengue virus envelope protein are found to interact with plasmin. These findings corrected earlier reports in the literature, which suggested that KR1-3 domains are involved in binding with domain III of the dengue virus envelope protein.
The research findings were published in the journal Protein Science.
Prof Mok said, “We are glad to have clarified inaccuracies in the literature. Our findings reveal new mechanisms of dengue virus pathogenesis, which could pave the way for innovative approaches to tackle vector-borne viruses.”
In the future, the group plans to study the interaction of other arboviruses, such as Zika and Chikungunya viruses, with plasmin. They also aim to determine the crystal structure of the protein complex formed between plasmin kringle-domains and the envelope protein of the dengue virus.
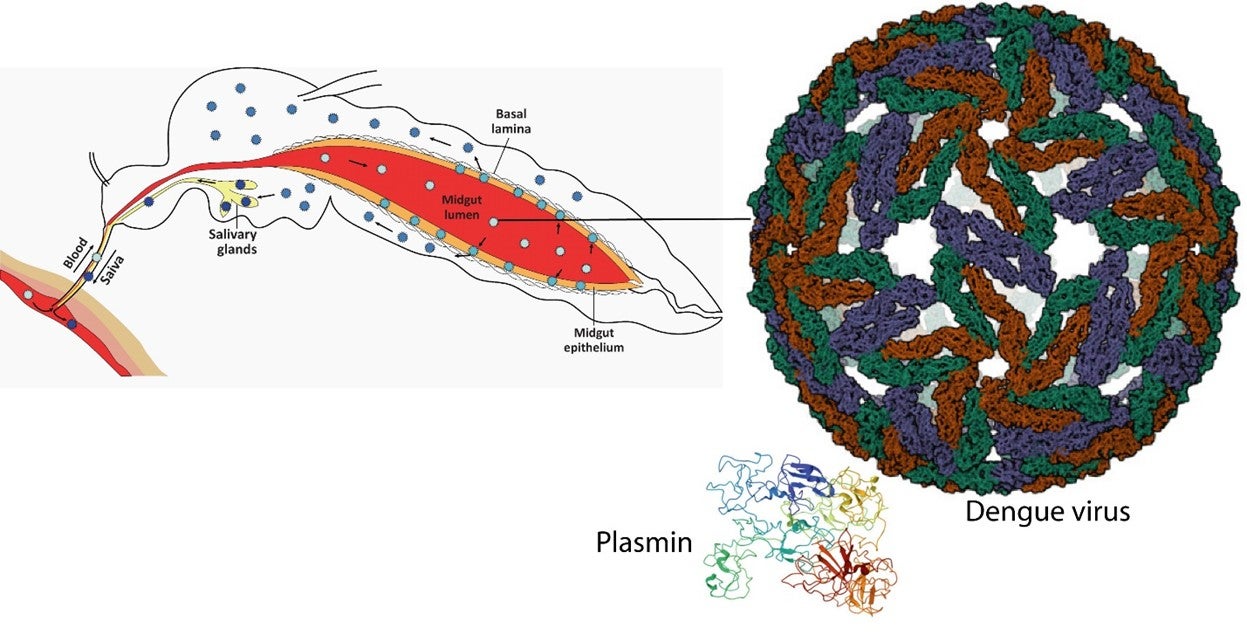
Diagram showing how the dengue virus captures plasmin using its envelope protein. The hijacked plasmin is used to enhance the permeability of the mosquito midgut to allow the dengue virus to escape and infect the salivary glands of the mosquito. Note that the diagram is not drawn to scale, and there will be more than one molecule of bound plasmin per viral particle. [Images of dengue virus and plasmin taken from Protein Data Bank https://www.rcsb.org, PDB ID: 3J35 and 4A5T, respectively. Diagram of mosquito taken from Ruckert, C. et al., 2018, Trends in Parasitology, 34, 310-321.]
Reference
YJ Yuen; T Sabitha; LJ Li; VA Walvekar; K Ramesh; RM Kini; J Sivaraman; YK Mok*, “Hijacking of plasminogen by dengue virus: The kringle-4 and -5 domains of plasminogen binds synergistically to the domain I of envelope protein” Protein Science 34(2):e70035 DOI: 10.1002/pro.70035 Published: 2025.


