Spatio-temporal dynamics of DNA replication
ZHAO Ziqing Winston (Group Leader, Chemistry) July 22, 2020NUS biophysical chemists have discovered a pattern that governs the propagation of deoxyribonucleic acid (DNA) replication in space and time by directly visualising the process in single human cells with sub-diffraction-limit resolution.
DNA replication is a process of crucial importance to the cell, and must be coordinated precisely to ensure that genomic information is duplicated once and only once during each cell cycle. It has long been known that mammalian DNA replication is initiated at numerous sites along the chromosomes known as replication origins, which are clustered into thousands of replication domains (RDs) across the genome. Such organisation is manifested in physical space as thousands of spatially clustered puncta within the cell nucleus. This is termed replication foci (RFi). Currently, how replication origins are chosen within individual RDs remains poorly understood, and it is unclear whether the origins are activated stochastically or preferentially near certain chromatin features.
A research team comprising Prof Ziqing Winston ZHAO from the Department of Chemistry and Centre for BioImaging Sciences, NUS, in collaboration with scientists from Peking University and University of Technology Sydney, has performed the first quantitative characterisation of the spatio-temporal organisation, morphology, and in situ epigenetic signatures of individual RFi in single human cells at super-resolution (~20 nm laterally). By combining the super-resolution imaging technique STORM with metabolic labelling and computational image segmentation analysis, the researchers found that DNA replication propagates by following a distinct, radially outward pattern during the early S-phase (DNA replication phase) of the cell cycle, but reverses its directionality to a radially inward propagation when it reaches late S-phase (see figure). Moreover, such pattern is diminished when there is a reduction in the expression of CTCF, a key nuclear protein that regulates 3-dimensional genome architecture. Together with simulation and bioinformatic analyses, the team’s findings point to a model in which replication is preferentially activated on looped chromatin structures organised by CTCF. This in turn suggests a non-random selection mechanism for replication activation at the sub-RD level, thereby providing strong evidence towards resolving a long-standing controversy.
Prof Zhao, who co-led the study said, “Our findings shed critical mechanistic insights into the role local epigenetic environment plays in coordinating DNA replication across the genome. More broadly, these findings could have fundamental implications for our understanding of how multi-scale chromatin architecture controls the organisation and dynamics of other intranuclear processes in space and time.”
The research team is planning to extend the approaches and framework developed in this study to explore other molecular factors implicated in the DNA replication process and the mechanistic correlation between replication and other chromatin-based transactions such as transcription.
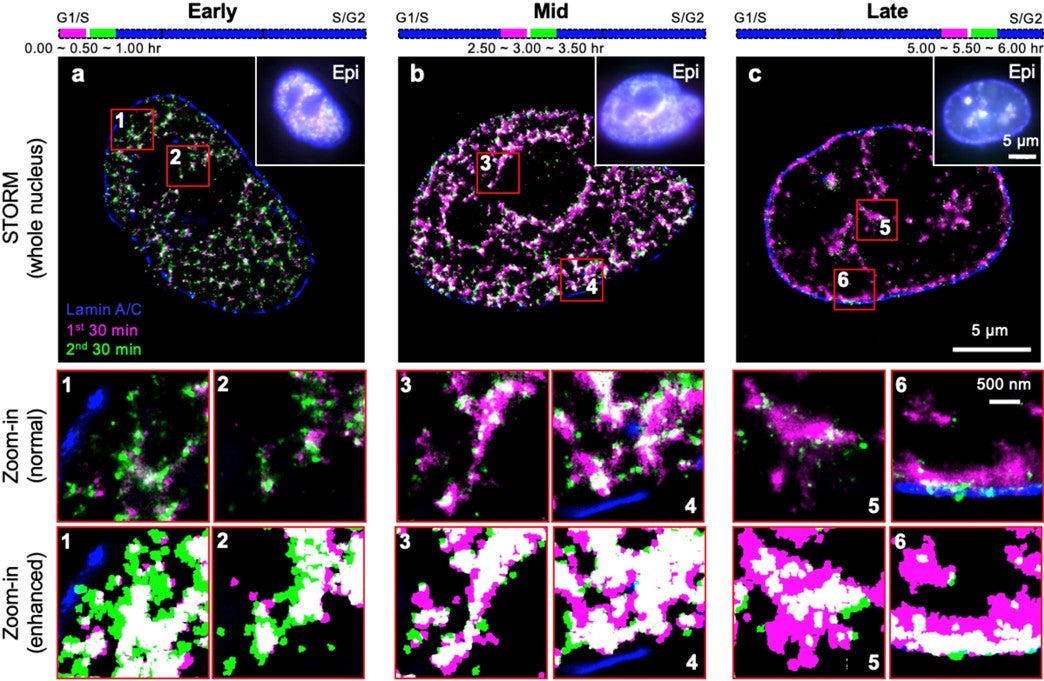
Figure shows the spatio-temporal patterns of RFi propagation dynamics at super-resolution across early (a), mid (b), and late (c) S-phase of the cell cycle in single human cells. Purple and green colours denote DNA replicated in two consecutive 30-min windows, respectively. Insets (numbered 1 through 6) show zoomed-in areas (in red boxes), displayed with both normal contrast (top) and enhanced contrast (bottom) between the two colours for better visualisation. [Credit: PNAS]
Reference
Su QP*; Zhao ZW*; Meng L; Ding M; Zhang W; Li Y; Liu M; Li R; GaoYQ; Xie XS*; Sun Y*, “Superresolution imaging reveals spatiotemporal propagation of human replication foci mediated by CTCF-organized chromatin structures” PROCEEDINGS OF THE NATIONAL ACADEMY OF SCIENCES OF THE UNITED STATES OF AMERICA DOI: 10.1073/pnas.2001521117 Published: 2020.


