Metabolic diversity of the “big six” of E. coli
YANG Hongshun (Group Leader, Food Science & Technology) April 13, 2020NUS food scientists have discovered that the six major strains of Escherichia coli (E. coli) that cause foodborne illnesses have different metabolisms and tolerance towards acidic conditions.
Pathogenic E. coli are responsible for a number of foodborne disease outbreaks. Among the hundreds of different E. coli serogroups (strains), pathogen E. coli O157:H7 is the most widely recognised due to the severity of the foodborne illnesses it causes. Apart from E. coli O157, there are another six serogroups that are identified by the United States Food and Drug Administration as emerging pathogens commonly found in foodborne disease outbreaks. These are known as the “big six”. As part of food safety, it is important to be able to characterise these pathogens and understand their behaviour so that proper measures can be developed to keep them at bay.
A research team led by Prof YANG Hongshun from the Department of Food Science and Technology, NUS used metabolomics technologies to study the adaptive response of pathogens under different inactivation stressors (electrolysed water, ultrasound, natural antibacterial agent, etc.). In their study, the team investigated the metabolic profiles of eight E. coli serotypes including E. coli O157:H7 and the “big six” using nuclear magnetic resonance (NMR). The results showed that metabolic diversity existed among the eight strains and different strains exhibited different tolerance towards acidic conditions.
The research team found that the pathogenic serotypes require higher energy production to fuel their physiological activities when compared to the non-pathogenic strain. The metabolic differences under acid stress indicate that energy and amino acid metabolisms (especially the glutamic acid dependent system) contributed to the different acidic adaptive capacities of the E. coli serotypes. These results suggest that the control of related metabolic pathways during sanitising treatments could potentially improve bacteria inactivation.
Traditional bacteria characterisations involve the physical examination of the morphological features using microscopes and biochemical characterisation of their biological properties using cultured bacteria on different media. These tests are very time-consuming, complex, and labour-intensive. In contrast, NMR provides a convenient, nondestructive and highly automated method of obtaining a comprehensive metabolic profile of the microorganisms.
Mr CHEN Lin, a Ph.D. student working on the project, said, “By monitoring the changes of microbial metabolomics, a precise snapshot of cells with different physicochemical states can be obtained. The research outcomes from the metabolomics analysis of the ‘big six’ pathogens provided a better understanding of food safety related bacterial properties.”
“These findings provide valuable references for the development of much effective food safety control measures, especially for controlling the emerging ‘big six’,” added Prof Yang.
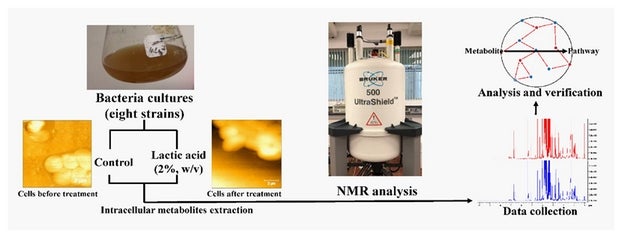
Figure shows the process involved in examining the acidic tolerance of E. coli strains using nuclear magnetic resonance (NMR). Bacteria cultures were prepared and separated into two sets, with one of them subject to acid treatment.
Reference
Chen L; Zhao X; Wu J; Liu Q; Pang X; Yang H*, “Metabolic characterisation of eight Escherichia coli strains including “Big Six” and acidic responses of selected strains revealed by NMR spectroscopy” FOOD MICROBIOLOGY Volume: 88 Article: 103399 DOI: 10.1016/j.fm.2019.103399 Published: 2020.


