Dynamic switch for FUS proteinopathy
July 18, 201718 Jul 2017. NUS biologists have discovered that dynamics of the FUS protein plays a key role in FUS proteinopathy that causes certain neurological diseases.
Amyotrophic lateral sclerosis (ALS) is a neurological disease that causes the death of nerve cells, which control voluntary muscle movements. Patients suffering from it gradually become weaker over time and have difficulties with their daily activities such as walking, breathing and talking. Despite its first discovery in 1869 by French neurologist Jean-Martin CHARCOT, the mechanism causing ALS is still a mystery and there is no effective treatment for this condition. ALS is a common neuromuscular disease worldwide. It affects people from different races and ethnic groups. This disease places a great burden on patients, their families and society. There is a campaign known as the ALS Ice Bucket Challenge to increase awareness of this disease.
A team led by Prof SONG Jianxing from the Department of Biological Sciences, NUS has succeeded in studying the FUS protein and its dissected domains, which naturally tend to form clusters. The FUS protein is known to be involved in the development of ALS and frontotemporal dementia (FTD). Their study showed that the FUS N- and C-terminal low-complexity (LC) domains are intrinsically disordered, and only the nucleic-acid recognition domain (RRM) is folded. However, RRM can spontaneously self-assemble into amyloid fibrils and it appears to play a crucial role in FUS proteinopathy. “The results shed light on a long-standing puzzle that FUS and TDP-43 cytotoxicity is mediated by their capacity in binding nucleic acids,” Prof Song commented.
Over the past years, Prof Song and his research team have been working on decoding structures, dynamics, self-assembly and membrane-interaction of a list of aggregation-prone proteins. This includes P56S-MSP, SOD1, TDP-43, FUS and E. coli ribosomal S1 proteins. They have been focusing on addressing an emerging and extremely challenging question of why aggregation-prone proteins can trigger an increasing spectrum of human diseases. Their work also involved cellular ageing down to E. coli cells. The studies established that there is a fine balance between the normal function and the potential to cause neurodegeneration, thus supporting the emerging idea that protein aggregation in neurologic disease may be an exaggeration of the normal functions of the aggregating proteins. Furthermore, membrane-interaction has been established as a common mechanism for aggregation-prone proteins to trigger human diseases and ageing. This is supported by their studies on “insoluble” proteins, which suggest that the driving forces for protein aggregation and interaction with membranes are at least overlapped, or even two sides of the same coin.
In the future, the research team plans to focus more on understanding why and how protein aggregation triggers cellular ageing and human diseases beyond neurodegenerative diseases.
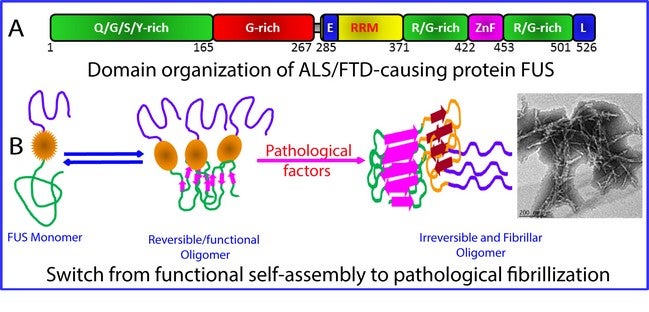
Figure shows the (A) domain structure of the 526-residue FUS protein and (B) model for the critical role of the RRM domain in exaggerating the reversible self-assembly underlying phase separation to irreversible fibrillisation characteristic of FUS proteinopathy including ALS and FTD. [Image credit: Song Jianxing]
- High-order secret decrypted for TDP-43 causing amyotrophic lateral sclerosis
- Protein dynamics determinant for ALS
References
1. Lu Y; Lim LZ; Song JX*, “RRM domain of ALS/FTD-causing FUS characteristic of irreversible unfolding spontaneously selfassembles into amyloid fibrils” SCIENTIFIC REPORTS Volume: 7 Article Number: 1043 DOI: 10.1038/s41598-017-01281-7 Published: 2017.
2. Lim LZ; Wei Y; Lu Y; Song JX*, “ALS-Causing Mutations Significantly Perturb the Self-Assembly and Interaction with Nucleic Acid of the Intrinsically Disordered Prion-Like Domain of TDP-43” PLOS BIOLOGY Volume: 14 Issue: 1 Article Number: e1002338 DOI: 10.1371/journal.pbio.1002338 Published: 2016.
3. Qin H; Lim LZ; Wei Y; Song JX*, “TDP-43 N terminus encodes a novel ubiquitin-like fold and its unfolded form in equilibrium that can be shifted by binding to ssDNA”PROCEEDINGS OF THE NATIONAL ACADEMY OF SCIENCES OF THE UNITED STATES OF AMERICA Volume: 111 Issue: 52 Pages: 18619-18624 DOI: 10.1073/pnas.1413994112 Published: 2014
4. Song JX*, “Insight into “insoluble proteins” with pure water” FEBS LETTERS Volume: 583 Issue: 6 Pages: 953-959 DOI: 10.1016/j.febslet.2009.02.022 Published: 2009 (Invited Review).
5. Song JX*, “Transforming Cytosolic Proteins into “Insoluble” and Membrane-toxic Forms Triggering Diseases/Aging by Genetic, Pathological or Environmental Factors”PROTEIN AND PEPTIDE LETTERS Volume: 24 Issue: 4 Pages: 294-306 DOI: 10.2174/0929866524666170209154001 Published: 2017 (Invited Review).


