Counting motifs in the human interactome
ZHANG Louxin ((Group Leader, Mathematics) ) November 17, 201617 Nov 2016. NUS mathematicians have developed an unbiased and consistent estimator for counting motif (pattern) of gene regulatory relationships.
Living cells are the products of the co-action of thousands of proteins. Some proteins, called transcription factors (TFs), serve to activate others. By binding to the so-called promoter region at the start of a target gene, a TF activates the gene, causing the cell to produce the corresponding protein. The relationship between TFs and target genes is modelled as a transcriptional regulatory network in biology (see the Figure below). The increasing availability of genomic and proteomic data has propelled network biology to the frontier of biomedical research.
Collaborating with Prof CHOI Kwok Pui, Prof ZHANG Louxin from Department of Mathematics, NUS has been working on the topological and dynamic properties of human cell-specific transcriptional regulatory networks. The feed-forward loop (FFL) (a type of connection pattern) and several other graphlets (called motifs) are found to be over-represented (having greater significance) in cellular protein networks. These motifs often represent functional units of biological processes in cells. As the network data (which are part of the true cellular protein networks) that have been reported by biological researchers contain a remarkably large number of incorrect protein relationships for humans, it is extremely challenging to estimate whether or not a motif is over- or under-represented for a given cellular protein network.
Taking link errors (incorrect biological relationships) into account, they developed an unbiased and consistent estimator for counting the motifs. This method has been found to work well for both undirected and directed networks. With the capability to account for link errors, this method greatly extends the previous work of Stumpf et al. (Proc. Natl Acad. Sci. USA, vol.105, 2008).
By applying this new method to 41 human cell-specific TF regulatory networks, they discovered that the TF regulatory network for embryonic stem cells has the smallest number of occurrences relative to its network size for FFL. They also found that 41 human cell-specific TF regulatory networks are significantly different.
The researchers are exploring the dynamic properties of cellular protein networks and using cellular networks to study the genetic variability of breast cancer.
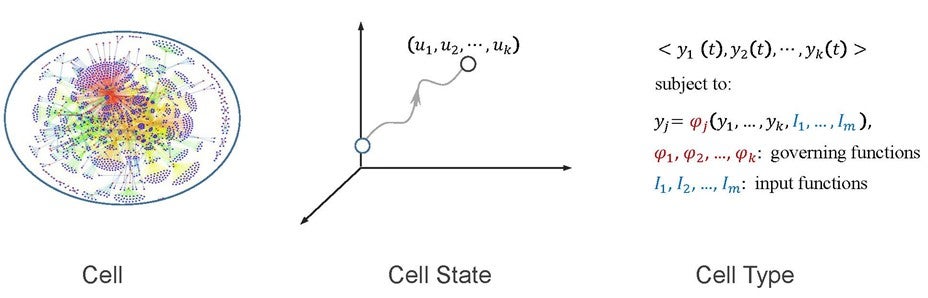
Figure shows that in network biology, a cell is modelled as a biological network, where dots are molecules, such as proteins and genes, and links represent some biological relationship between molecules. A cell state is a numerical vector, in which each module has a value. A cell type becomes a dynamic system specified by a set of governing and input functions.
References
1. Tran NH; Choi KP; Zhang LX*, “Counting motifs in the human interactome” NATURE COMMUNICATIONS Volume: 4 Article Number: 2241 DOI: 10.1038/ncomms3241 Published: 2013
2. Zhang SH; Tian DC; Tran NH; Choi KP*; Zhang LX*, “Profiling the transcription factor regulatory networks of human cell types” NUCLEIC ACIDS RESEARCH Volume: 42 Issue: 20 Pages: 12380-12387 DOI: 10.1093/nar/gku923 Published: 2014


