Arresting cancer by energy starvation
Jayaraman SIVARAMAN ((Group Leader, Biological Sciences) ) April 18, 201718 Apr 2017. NUS biologists have discovered a medicinal compound that can potentially prevent cancer cells from becoming a tumour.
Cancer, one of the top killer diseases, occur when cell growth goes out of control. It then invades other healthy tissues and spreads to other parts of the body. NUS scientists have discovered a compound which may stop its progression. They have found out how this compound, that is undergoing pre-clinical trials as a potential drug, can “starve” cancer cells of energy. Cancer cells need a lot of energy to grow and develop. Cutting off its energy source prevents the cancer cells from developing into a tumour.
A team led by Prof Jayaraman SIVARAMAN from the Department of Biological Sciences (DBS), in collaboration with Prof LOW Boon Chuan from DBS and the Mechanobiology Institute at NUS, has unveiled the structure and function of glutaminase, a key enzyme in cancer metabolism. They have also characterised the interaction between glutaminase and the chemical inhibitors, BPTES (bis-2-(5- phenylacetamido-1, 2, 4-thiadiazol- 2-yl) ethyl sulfide) and its derivatives [1,3] along with the inhibition of the ERK signalling pathway.
The kidney-type glutaminase (KGA) is a key enzyme responsible for the process of glutaminolysis, which is harnessed by cancer cells (the Warburg Effect) to thrive and become a tumour. The enzyme glutaminase catalyses the first and most crucial step in glutamine metabolism. Many cancer cells cannot survive without glutamine – referred to as glutamine addiction. However, nothing was known about the interactions between KGA and its inhibitor, BPTES.
The investigators, in collaboration with Prof Valiyaveettil SURESH from the Department of Chemistry, NUS, and colleagues from the Karolinska Institute, Sweden, determined the mechanism by which BPTES/derivatives binds to, and inhibits KGA, abolishing the cancer cells’ energy source and thus potentially blocking their growth. Most strikingly, their study revealed that BPTES/derivatives inhibit KGA by inducing a dramatic allosteric conformational change. This result will aid in the development of better small-molecule inhibitors and analogues of BPTES that target glutaminase activity with reduced toxicity. The research team also concluded that a combination of BPTES and another inhibitor of a glutaminase-activating component (ERK signalling pathway) would be even more effective at reducing glutaminase activity and cell growth [1,2,3].
These findings could offer a more potent but less cytotoxic cancer treatment regime and show promise for a new dual-drug cancer treatment that may be more effective and with fewer side effects, especially for patients with cancers such as lymphoma, prostate cancer, glioblastoma, breast cancer and kidney cancer.
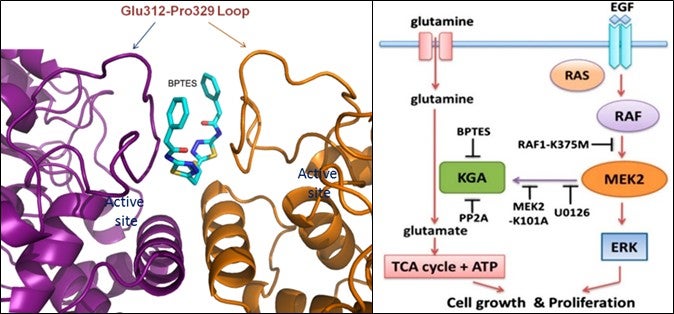
(Left) Figure 1 shows a close-up view of the BPTES binding pocket on the surface exposed region of the loop Glu312-Pro329 at the KGA dimer interface. (Right) Figure 2 shows the schematic model depicting the synergistic cross-talk between KGA-mediated glutaminolysis and EGF-activated Raf-Mek-Erk signalling. [Image credit: THANGAVELU K, Sarath RAMACHANDRAN and J. Sivaraman].
References
1. Ramachandran S; Pan CQ; Zimmermann SC; Duvall B; Tsukamoto T; Low BC; Sivaraman J*, “Structural basis for exploring the allosteric inhibition of human kidney type glutaminase” ONCOTARGET Volume: 7 Issue: 36 Pages: 57943-57954 DOI: 10.18632/oncotarget.10791 Published: 2016.
2. Thangavelu K; Chong QY; Low BC; Sivaraman J*, “Structural basis for the active site inhibition mechanism of human kidney-type glutaminase (KGA)” SCIENTIFIC REPORTS Volume: 4 Article Number: 3827 DOI: 10.1038/srep03827 Published: 2014.
3. Thangavelu K; Pan CQ; Karlberg T; Balaji G; Uttamchandani M; Suresh V; Schüler H; Low BC*; Sivaraman J*, “Structural basis for the allosteric inhibitory mechanism of human kidney-type glutaminase (KGA) and its regulation by Raf-Mek-Erk signaling in cancer cell metabolism” PROCEEDINGS OF THE NATIONAL ACADEMY OF SCIENCES OF THE UNITED STATES OF AMERICA Volume: 109 Issue: 20 Pages: 7705-7710 DOI: 10.1073/pnas.1116573109 Published: 2012.


