Mutation patterns in DNA sequences
August 03, 20173 Aug 2017. NUS statistician has developed new statistical models that capture specific patterns within the DNA sequence which seem to influence genetic mutation.
Around half a century ago, striking similarities in the biological sequences from different species were first observed in amino acids and then in the hereditary molecule, DNA. For example, the cellular factory of proteins, known as the ribosome, is similar across plants and animals. Collectively, such findings provide strong evidence to support the existence of a common origin of life. The extent of molecular similarity between two species is a reflection of how closely they are related in their evolutionary process. The similarities can also be displayed through other attributes, such as physiological forms and functions. Statistical models of evolution can be used to learn about the rates of evolution, by analysing and comparing certain patterns found in the DNA sequences of plant and animal species.
Prof YAP Von Bing from the Department of Statistics and Applied Probability, NUS has developed statistical models to describe the evolutionary change in DNA sequences. This work is in collaboration with Prof Gavin HUTTLEY and his research group at the Australian National University. An important goal of their research programme is the quantification of the magnitude of various evolutionary forces, such as spontaneous mutation, DNA repair mechanisms, and natural or artificial selection. Techniques developed by the team can be applied towards the measurement of evolutionary distances and reconstruction of the evolutionary relationships between different organisms, providing new insights on the understanding of human health and disease.
The DNA molecule consists of a long chain of chemical units known as bases, of which there are four kinds: A, C, G and T. Embedded in the DNA are instructions for the growth, development, functioning and reproduction of organisms. The simplest mechanism of DNA change is point substitution: the replacement of a base by another one, say C to T. The substitution process may depend on the identity of the neighbouring bases, which are known as the context. The most famous example is the CpG effect: whenever C is followed by G in the DNA chain, there is a much higher (about 10 times) chance that C is substituted by T, than if the neighbouring base were a non-G. The research group made a first attempt at a systematic detection of contexts that seem to affect point mutation rates in human germline and malignant melanoma cells. Using a log-linear model for substitution rates, they have recovered the CpG effect and discovered some other new influencing factors.
Prof Yap said, “Such context-dependent mutation events are important for understanding the cellular bases for evolution, and may have major implications for understanding genetic diseases, including cancers.”
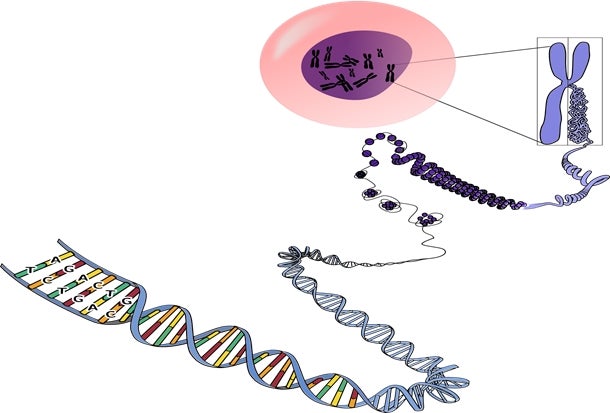
Figure show that the DNA molecule which is made up of bases A,C,G and T, encodes the set of genetic instructions and codes that determine the function of a cell. [Image credit: https://pixabay.com/en/genetics-chromosomes-rna-dna-156404/]
Reference
Zhu Y; Neeman T; Yap VB; Huttley G, “Statistical methods for identifying sequence motifs affecting point mutations” GENETICS Volume: 205 Issue: 2 Pages: 843-856 DOI: 10.1534/genetics.116.195677 Published: 2017.


