Protein extraction from green algae
LIN Qingsong (Group Leader, Biological Sciences) () August 25, 2016NUS biologists have developed an effective method for protein extraction from green algae to detect environmental pollutants.
25 Aug 2016. Pesticides such as Cypermethrin (CYP) are widely used in large-scale commercial agricultural applications but their residues may have an impact on the ecosystem. Proteomics analysis is a technique which could provide toxicity assessment information on the adverse effects of these pollutants at an early stage when organisms such as algae are exposed at low dosage. Samples containing the proteins are from the organisms to perform the analysis.
A research team led by Prof LI Fong Yau, Sam from the Department of Chemistry and Dr LIN Qingsong from the Department of Biological Sciences at NUS discovered that protein extraction methods involving a chemical compound, phenol, produced more accurate results compared to other conventional methods of algae proteome analysis. Knowing that trichloroacetic acid can precipitate protein and inhibit protease activity effectively, they combined it with the use of phenol for protein extraction. This new method reduced the exposure time of protein samples in an acidic environment, which helped to reduce protein degradation and improve protein yield. Their study provided insights on protein extraction efficiency in terms of the number of identifiable proteins and the quantity which can be reliably extracted using the different methods.
Although algae proteomics has wide applications in various fields such as biofuels, biomonitoring and pollution control, current findings on the evaluation of different protein extraction methods are mainly based on plant tissues. There is limited information on the optimal method for algae species. This study has helped improve understanding on protein extraction for algae proteome profiling research.
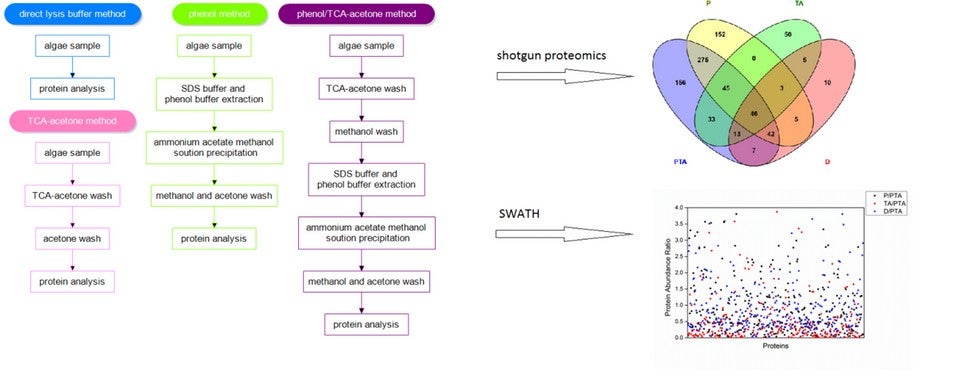
Figure shows the protocols of four protein extraction methods and the results of proteomics investigation. The Venn diagramme shows identified proteins from the four different extraction methods. The bottom right figure shows ratios of protein abundance from the other three methods compared with the optimal one. PTA: Phenol/TCA-acetone method; P: Phenol method; TA: TCA-acetone method; D: Direct lysis buffer method.
Reference
Gao Y., Lim T.K., Lin Q., Li S.F. “Identification of cypermethrin induced protein changes in green algae by iTRAQ quantitative proteomics”. J. Proteomics, 139:67-76 (2016).
Wang W., Vignani R., Scali M., Crest M. “A universal and rapid protocol for protein extraction from recalcitrant plant tissues for proteomics analysis”. Electrophoresis, 27: 2782 (2006).


